New Carnegie Mellon Dynamic Statistical Model Follows Gene Expressions Over Time
Future Applications Could Range From fMRI Data to Social Networks and More
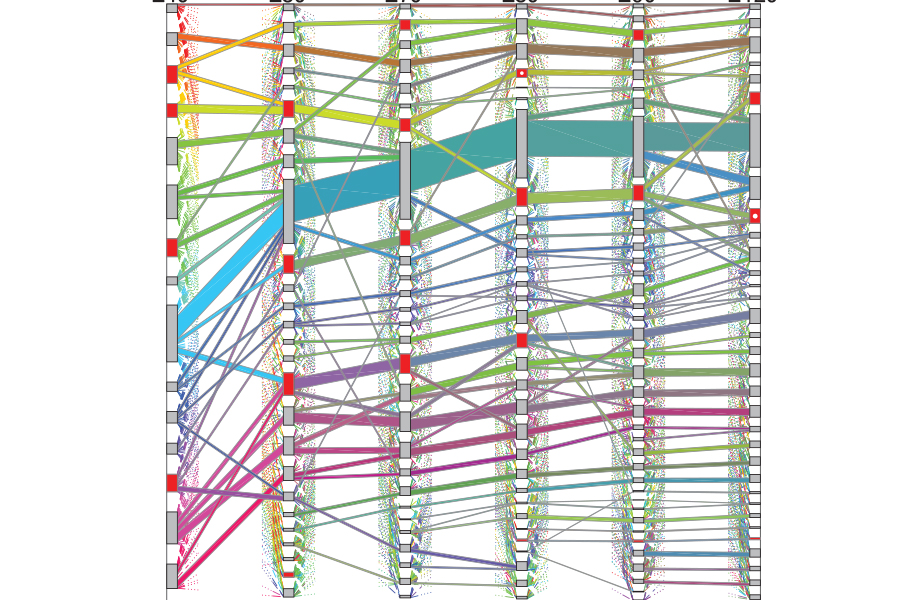 Researchers at Carnegie Mellon University have developed a new dynamic statistical model to visualize changing patterns in networks, including gene expression during developmental periods of the brain.
Researchers at Carnegie Mellon University have developed a new dynamic statistical model to visualize changing patterns in networks, including gene expression during developmental periods of the brain.
Published in the Proceedings of the National Academy of Sciences, the model now gives researchers a tool that extends past observing static networks at a single snapshot in time, which is hugely beneficial since network data are usually dynamic. The analysis of network data—or the study of relationships from a large-scale view—is an emerging field of statistics and data science.
“For any dataset with a dynamic component, people can now use this in a powerful way to find communities that persist and change over time,” said Kathryn Roeder, the UPMC Professor of Statistics and Life Sciences in the Dietrich College of Humanities and Social Sciences. “This will be very helpful in understanding how certain diseases and disorders progress. For example, we know that certain genes are responsible for autism and can use our model to give us insight into at what point the disorder begins developing.”
The model, Persistent Communities by Eigenvector Smoothing (PisCES), combines information across a series of networks, longitudinally, to strengthen the inference for each period. The CMU team used PisCES to follow neural gene expressions from conception through adulthood in rhesus monkey brains to find out what genes work together during different points of development.
“Our visualization method combines two different existing tools: community detection, which is a popular tool for static network data, and sankey plots, which are often used to visualize ‘flows’ of information. Our visualization organizes the actors of the network into communities that evolve over time and then depicts the evolving community memberships as a series of flows between the communities,” said David Choi, assistant professor of statistics and information systems at the Heinz College of Information Systems and Public Policy.
Their analysis revealed the existence of change points as well as periods of persistent gene community structure, including a dynamic community of genes involved in neural projection guidance that was highly active during the mid to late fetal period. This particular community includes many genes associated with risk for autism.
“Essentially our goal was to add ‘smoothing’ to community detection to eliminate the ‘noise,’ and we were able to do that,” Choi said.
Although the team piloted the model by visualizing the changing patterns in the ways that genes work together, the hope is that the method could be applied to social networks, dynamic diffusion networks in physics and other relational situations.
“The model is really flexible, and we are already starting to use it with fMRI data to understand how regions of the brain interconnect and change over time,” said Fuchen Liu, a Ph.D. student in the Department of Statistics and Data Science.
Developing a new dynamic statistical model to follow neural gene expressions over time is one of the many brain research breakthroughs to happen at Carnegie Mellon. CMU has created some of the first cognitive tutors, helped to develop the Jeopardy-winning Watson, founded a groundbreaking doctoral program in neural computation, and is the birthplace of artificial intelligence and cognitive psychology. Building on its strengths in biology, computer science, psychology, statistics and engineering, CMU launched BrainHub, an initiative that focuses on how the structure and activity of the brain give rise to complex behaviors.
This work was funded by the National Institutes of Mental Health and Simons Foundation.
Original Article By: Shilo Rea